|
MCTDH-X
v2.22
|
|
MCTDH-X
v2.22
|
This module collects the routines for the analysis of two-body quantities in R-MCTDHB simulations. More...
Public Member Functions | |
| subroutine | get_correlationsskewdiagonal1d (time, PSI, mode, rho_jk, rho_ijkl, dilation) |
| This routine computes the skew diagonal of 1D correlation functions G1(x,x'=-x), and G2(x,x'=-x), and writes them to a file. The first four columns contain x,x'. More... | |
| subroutine | get_2bentropy (FT_PSI, PSI, rho_ijkl, TIME) |
| Compute the entropy of the two-body density diagonal in real and in momentum space. Compute the entropy of the geminal occupations. More... | |
| subroutine | diag_geminal (Time, NatGemOcc, PSI, jobtype, InEntropy) |
| Natural geminals' analysis: Diagonalize reduced two-body density matrix elements and return eigenvalues (Natural Geminal Occupations). More... | |
| subroutine | lossops_two_bosons (time, PSI, rho_ijkl, border) |
| This routine computes the expectation values for N=2 particle loss operators when partitioning the two particle Hilbert space at 'border'. More... | |
| subroutine | get_corr_one_restricted (time, PSI, mode, rho_jk, xini, xfin, xpts, kxini, kxfin, kpts) |
| This routine computes the correlation functions on a restricted subspace from x(k)ini to x(k)fin with x(k)pts points. More... | |
| subroutine | get_corr_two_restricted (time, PSI, mode, rho_jk, rho_ijkl, xini, xfin, xpts, kxini, kxfin, kpts) |
| This routine computes the correlation functions G2(r,r',r',r), where r=(x/k) in a restricted subspace from x(k)ini to x(k)fin with x(k)pts. More... | |
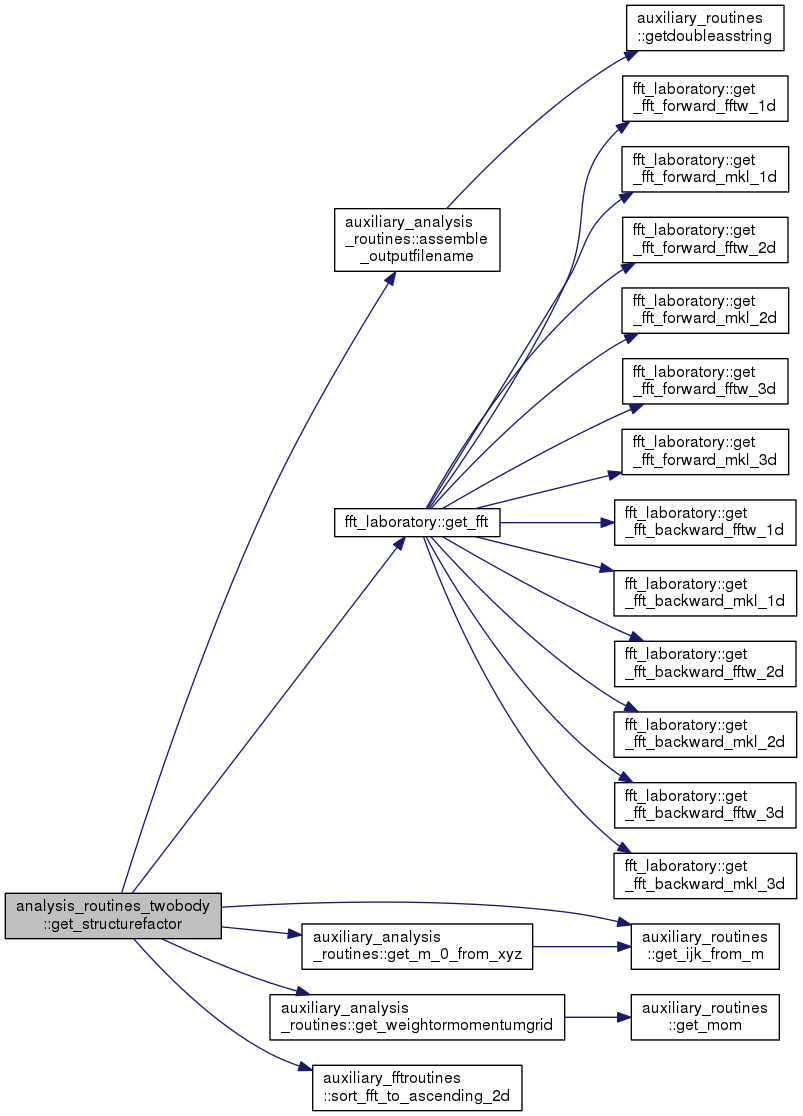
| subroutine | get_structurefactor (time, PSI, rho_jk, rho_ijkl, xref, yref, zref) |
| subroutine | get_correlations (time, PSI, mode, rho_jk, rho_ijkl, Dilation, CorrCoef) |
| This routine computes the correlation functions G1(r,r), G1(r,r'), G1(r',r') and G2(r,r',r',r), where r=(x,y,z), and writes them to a file. The first six columns contain x,y,z,x',y',z'. More... | |
| subroutine | get_correlations_nlevel (time, PSI, mode, rho_jk, rho_ijkl, dilation) |
| This routine computes the correlation functions G1(r,r), G1(r,r'), G1(r',r') and G2(r,r',r',r), where r=(x,y,z), and writes them to a file. The first six columns contain x,y,z,x',y',z'. More... | |
This module collects the routines for the analysis of two-body quantities in R-MCTDHB simulations.
| subroutine analysis_routines_twobody::diag_geminal | ( | real*8 | Time, |
| real*8, dimension(morb*(morb+1)/2) | NatGemOcc, | ||
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| integer | jobtype, | ||
| logical, optional | InEntropy | ||
| ) |
Natural geminals' analysis: Diagonalize reduced two-body density matrix elements and return eigenvalues (Natural Geminal Occupations).
References auxiliary_analysis_routines::assemble_outputfilename(), and auxiliary_analysis_routines::get_full_rijkl().
Referenced by analysis_core::apply_analysis(), and get_2bentropy().

| subroutine analysis_routines_twobody::get_2bentropy | ( | complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | FT_PSI, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| complex*16, dimension(morb,morb,morb,morb) | rho_ijkl, | ||
| real*8 | TIME | ||
| ) |
Compute the entropy of the two-body density diagonal in real and in momentum space. Compute the entropy of the geminal occupations.
Many-Body entropy related to the natural geminal occupations.
Write different entropies to a file.
References diag_geminal(), auxiliary_routines::get_ijk_from_m(), and auxiliary_analysis_routines::get_weightormomentumgrid().
Referenced by analysis_core::apply_analysis().

| subroutine analysis_routines_twobody::get_corr_one_restricted | ( | real*8, intent(in) | time, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| integer, intent(in) | mode, | ||
| complex*16, dimension(morb,morb), intent(in) | rho_jk, | ||
| real*8 | xini, | ||
| real*8 | xfin, | ||
| integer, intent(in) | xpts, | ||
| real*8 | kxini, | ||
| real*8 | kxfin, | ||
| integer, intent(in) | kpts | ||
| ) |
This routine computes the correlation functions on a restricted subspace from x(k)ini to x(k)fin with x(k)pts points.
References auxiliary_analysis_routines::assemble_outputfilename(), auxiliary_routines::get_ijk_from_m(), and auxiliary_routines::get_mom().
Referenced by analysis_core::apply_analysis().

| subroutine analysis_routines_twobody::get_corr_two_restricted | ( | real*8, intent(in) | time, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| integer, intent(in) | mode, | ||
| complex*16, dimension(morb,morb), intent(in) | rho_jk, | ||
| complex*16, dimension(morb,morb,morb,morb), intent(in) | rho_ijkl, | ||
| real*8 | xini, | ||
| real*8 | xfin, | ||
| integer, intent(in) | xpts, | ||
| real*8 | kxini, | ||
| real*8 | kxfin, | ||
| integer, intent(in) | kpts | ||
| ) |
This routine computes the correlation functions G2(r,r',r',r), where r=(x/k) in a restricted subspace from x(k)ini to x(k)fin with x(k)pts.
References auxiliary_analysis_routines::assemble_outputfilename(), auxiliary_routines::get_ijk_from_m(), and auxiliary_routines::get_mom().
Referenced by analysis_core::apply_analysis().


| subroutine analysis_routines_twobody::get_correlations | ( | real*8, intent(in) | time, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| integer, intent(in) | mode, | ||
| complex*16, dimension(morb,morb), intent(in) | rho_jk, | ||
| complex*16, dimension(morb,morb,morb,morb), intent(in) | rho_ijkl, | ||
| integer, intent(in) | Dilation, | ||
| logical, optional | CorrCoef | ||
| ) |
This routine computes the correlation functions G1(r,r), G1(r,r'), G1(r',r') and G2(r,r',r',r), where r=(x,y,z), and writes them to a file. The first six columns contain x,y,z,x',y',z'.
References auxiliary_analysis_routines::assemble_outputfilename(), auxiliary_routines::get_ijk_from_m(), and auxiliary_analysis_routines::get_weightormomentumgrid().
Referenced by analysis_core::apply_analysis().

| subroutine analysis_routines_twobody::get_correlations_nlevel | ( | real*8, intent(in) | time, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb,nlevel) | PSI, | ||
| integer, intent(in) | mode, | ||
| complex*16, dimension(morb,morb), intent(in) | rho_jk, | ||
| complex*16, dimension(morb,morb,morb,morb), intent(in) | rho_ijkl, | ||
| integer | dilation | ||
| ) |
This routine computes the correlation functions G1(r,r), G1(r,r'), G1(r',r') and G2(r,r',r',r), where r=(x,y,z), and writes them to a file. The first six columns contain x,y,z,x',y',z'.
References auxiliary_analysis_routines::assemble_outputfilename(), auxiliary_routines::get_ijk_from_m(), and auxiliary_analysis_routines::get_weightormomentumgrid().
Referenced by analysis_core::apply_analysis().

| subroutine analysis_routines_twobody::get_correlationsskewdiagonal1d | ( | real*8, intent(in) | time, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| integer, intent(in) | mode, | ||
| complex*16, dimension(morb,morb), intent(in) | rho_jk, | ||
| complex*16, dimension(morb,morb,morb,morb), intent(in) | rho_ijkl, | ||
| integer, intent(in) | dilation | ||
| ) |
This routine computes the skew diagonal of 1D correlation functions G1(x,x'=-x), and G2(x,x'=-x), and writes them to a file. The first four columns contain x,x'.
References auxiliary_analysis_routines::assemble_outputfilename(), auxiliary_routines::get_ijk_from_m(), auxiliary_analysis_routines::get_weightormomentumgrid(), and auxiliary_routines::getdoubleasstring().
Referenced by analysis_core::apply_analysis().

| subroutine analysis_routines_twobody::get_structurefactor | ( | real*8, intent(in) | time, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| complex*16, dimension(morb,morb), intent(in) | rho_jk, | ||
| complex*16, dimension(morb,morb,morb,morb), intent(in) | rho_ijkl, | ||
| real*8, intent(in) | xref, | ||
| real*8, intent(in) | yref, | ||
| real*8, intent(in) | zref | ||
| ) |
References auxiliary_analysis_routines::assemble_outputfilename(), fft_laboratory::get_fft(), auxiliary_routines::get_ijk_from_m(), auxiliary_analysis_routines::get_m_0_from_xyz(), auxiliary_analysis_routines::get_weightormomentumgrid(), and auxiliary_fftroutines::sort_fft_to_ascending_2d().
Referenced by analysis_core::apply_analysis().

| subroutine analysis_routines_twobody::lossops_two_bosons | ( | real*8, intent(in) | time, |
| complex*16, dimension(ndvr_x*ndvr_y*ndvr_z,morb) | PSI, | ||
| complex*16, dimension(morb,morb,morb,morb), intent(in) | rho_ijkl, | ||
| real*8, intent(in) | border | ||
| ) |
This routine computes the expectation values for N=2 particle loss operators when partitioning the two particle Hilbert space at 'border'.
References auxiliary_routines::get_ijk_from_m(), and auxiliary_routines::getdoubleasstring().
Referenced by analysis_core::apply_analysis().